Screening the Biomarkers and Related Medicines for Gastric Adenocarcinoma
By Juanjuan Lin1, Ning Xu2, Weifang An3Affiliations
doi: 10.29271/jcpsp.2024.03.290ABSTRACT
Objective: To search for potential biomarkers and available medicines for gastric adenocarcinoma.
Study Design: Experimental study.
Place and Duration of the Study: Scientific Research Section, Shenzhen Longhua District Central Hospital, Shenzhen, China, from January to April 2023.
Methodology: Datasets were retrieved from the Gene Expression Omnibus (GEO). Differential gene expression analysis between gastric adenocarcinoma and normal samples was conducted using GEO2R. Subsequent Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed via the Enrichr website. Protein-protein interaction (PPI) networks were established using the STRING website. The central hub genes were identified using the cytoHubba plugin integrated within Cytoscape. Finally, the GEPIA2 and QuartataWeb websites were employed to validate the expression levels of the hub genes and to identify potential medicines for gastric adenocarcinoma.
Results: In total, 133 DEGs were identified. GO analysis revealed that these DEGs predominantly participate in processes such as cell adhesion, positive regulation of cell proliferation, and extracellular matrix organisation. In the KEGG pathways, DEGs were significantly enriched in gastric acid secretion, protein digestion and absorption, and ECM-receptor interaction. Following the construction of the PPI network, 10 central hub genes were identified and validated using GEPIA2. Notably, among these hub genes, SERPINE1 demonstrated a significant association with the prognosis of gastric adenocarcinoma, and potential therapeutic agents were subsequently predicted.
Conclusion: SERPINE1 and potential therapeutic agents hold promise to enhance personalised diagnosis and treatment for gastric adenocarcinoma patients in the future.
Key Words: Biomarkers, Gastric adenocarcinoma, Bioinformatics, Differentially Expressed Genes (DEGs).
INTRODUCTION
Gastric carcinoma, often referred to as stomach cancer, is a malignant neoplasm originating from the epithelium of the gastric mucosa.1 Its pathogenesis is multifactorial and includes factors such as chronic gastritis, infection with Helicobacter pylori, specific dietary habits (e.g., high salt intake and consumption of preserved foods), tobacco use, and genetic predispositions.2 Gastric adenocarcinoma, a subtype that arises from the glandular cells of the gastric mucosa, represents approximately 95% of all stomach cancer cases.1 Thus, global incidence rates for gastric adenocarcinoma closely align with those for stomach cancer in general.
It is estimated that nearly one million new cancer cases diagnosed each year are of this subtype. Consistent with its prevalence, mortality rates for gastric adenocarcinoma are reflective of the broader category of stomach cancers. Specifically, of the roughly 723,000 annual stomach cancer-related deaths, a significant majority are due to adenocarcinoma.2
While there exist multiple methods for detecting gastric adenocarcinoma, they present certain limitations. Endoscopy, while effective, but its broad application is limited due to invasive nature, the cost implication, and reliant on the endoscopist’s skill. Tissue biopsies, though informative, carry risks like bleeding or infection, and might not always conclusively identify cancer. CT scans subject patients to radiation, and difficult to distinguish normal tissue, such as fat and muscles, from tumours. Tumour marker tests, such as CEA and CA19-9, should not be used in isolation for diagnosis, serving merely as references.3-5 Thus, there is a pressing need for the development of early and reliable detection methods for this significant health issue.
Bioinformatics is an interdisciplinary domain that melds insights from biology, computer science, mathematics, and statistics to aggregate, process, interpret, and analyse extensive biological datasets. This field arose from the imperative to manage and scrutinise the burgeoning volume of biological data, particularly from high-throughput endeavours such as genomic sequencing and proteomics.5-7 Offering potent tools for contemporary biology and medical research, bioinformatics profoundly influences areas like disease mechanism comprehension, medicine discovery, and personalised medicine. Numerous public databases provide clinical and sequencing data for download and analysis. By reanalysing these datasets, there is potential to unearth fresh perspectives on gastric adenocarcinoma research. While various studies have identified genes linked to gastric adenocarcinoma,8,9 a solitary gene cannot elucidate all underlying mechanisms. Thus, employing bioinformatics to uncover additional molecular biomarkers is pivotal for the early diagnosis and targeted therapy of gastric adenocarcinoma.
The objective of the study was to validate the expression levels of the hub genes and to identify potential medicines for gastric adenocarcinoma.
METHODOLOGY
This bioinformatics study was conducted from January to April 2023 at the Scientific Research Section, Shenzhen Longhua District Central Hospital, Shenzhen, China.
GSE118916, GSE103236, and GSE26942, were downloaded from GEO database. GSE26942 comprised 12 cancers and 205 normal samples. GSE118916 had 15 cancers and 15 normal tissues. GSE103236 comprised 9 cancers and 10 normal samples.
GEO2R online tool was utilised to identify differentially expressed genes (DEGs) between gastric adenocarcinoma and normal samples. Subsequently, Venn software was employed to identify common DEGs across three datasets. The cut-off criteria were set at |logFC| >2 and p <0.05. A logFC value less than -2 indicated down-regulation, whereas a value greater than 2 indicated up-regulation.
GO analysis was conducted to assess the cellular components, molecular functions, and enriched biological pathways associated with various genes and genomes.10 The KEGG pathway, a subsection of the KEGG database, was used for delineating molecular interactions and reaction networks across various biological processes with comprehensive graphical representations, facilitating an intuitive comprehension of distinct biological pathways, diseases, or functions as well as the pathways for upregulation or downregulation.11 Visualisation of DEGs cluster for biological process, cellular component, molecular function, and KEGG pathways (p <0.05) was achieved using the Enrichr website.
In order to find interaction of DEGs, online bioinformatic tool STRING was used. Then, the authors analysed and visualised PPI network of DEGs by using the Cytoscape software.12 The top ten hub genes from the PPI network were selected by MCC algorithm in the plugin cytoHubba (scores >2).13
GEPIA2 (Gene Expression Profiling Interactive Analysis 2) was used for analysing cancer gene expression based on RNA sequencing data. The tool primarily draws data from TCGA (The Cancer Genome Atlas) and GTEx (Genotype-Tissue Expression) projects, offering rapid and customisable functionalities that allow researchers to analyse gene expression differences between cancer samples and normal tissue samples.14 The hub genes' expression levels between 408 gastric adenocarcinoma and 211 normal samples were performed by using boxplots. Kaplan-Meier curves were conducted to evaluate the prognosis of gastric adenocarcinoma patients whose hub genes had been expressed.
Using QuartataWeb database, information for medicine-gene interaction was obtained based on over thirty reliable sources. Hub genes were inserted into the database, to screen compounds or medicines. Statistical significance was determined by applying t-test with p <0.05 taken as statistically significant.
RESULTS
DEGs (1749 in GSE103236, 419 in GSE26942, and 1851 in GSE118916) were identified using GEO2R. According to the Venn diagram, the overlap in the 3 datasets contained 133 genes. Upregulation of 30 genes and downregulation of 103 genes were observed.
In biological process (BP) annotation, it was mostly enriched in cell adhesion, positive regulation of cell proliferation, extracellular matrix organisation, positive regulation of cell migration, and positive regulation of extracellular-regulated kinase 1/2(ERK1/2) cascade. In cellular component (CC) annotation, it was enriched in plasma membrane, extracellular exosome, extracellular region, and extracellular space. In molecular function (MF) annotation, the DEGs were clustered in structural constituent of cytoskeleton, protease binding, WW domain binding, and extracellular matrix binding (Table I). Additionally, in KEGG pathway, the DEGs were clustered into ECM-receptor interaction, Chemical carcinogenesis, Metabolism of xenobiotics by cytochrome P450, and Complement and coagulation cascades (Table I).
As shown in Figure 1 (A and B), the red nodes represented up-regulated genes while the blue ones represented down-regulated genes. 103 genes/nodes with 160 edges enriched in PPI network. As shown in Figure 1B, the top 10 hub genes were: SPP1, SERPINE1, TIMP1, MMP3, BGN, SST, THBS2, GHRL, CHGA, and ATP4A.
To further identify the hub genes' reliability in gastric adenocarcinoma, the authors entered the hub genes into the Gene Expression Profiling Interactive Analysis (GEPIA2) website (http://gepia2.cancer-pku.cn/) to match the expression levels and prognostic information.
Table I: GO analysis and KEGG pathway analysis.
|
Category |
Term |
Count |
Ratio (%) |
p-value |
|
BP |
GO:0007155~cell adhesion |
13 |
9.774436 |
1.63E-04 |
|
BP |
GO:0008284~positive regulation of cell proliferation |
9 |
6.766917 |
0.023258 |
|
BP |
GO:0030198~extracellular matrix organisation |
9 |
6.766917 |
1.10E-04 |
|
BP |
GO:0010628~positive regulation of gene expression |
7 |
5.263158 |
0.013679 |
|
BP |
GO:0030335~positive regulation of cell migration |
6 |
4.511278 |
0.012201 |
|
BP |
GO:0034220~ion transmembrane transport |
6 |
4.511278 |
0.020454 |
|
BP |
GO:0070374~positive regulation of ERK1 and ERK2 cascade |
6 |
4.511278 |
0.009977 |
|
BP |
GO:0007586~digestion |
6 |
4.511278 |
1.05E-04 |
|
BP |
GO:0007596~blood coagulation |
5 |
3.759398 |
0.048729 |
|
BP |
GO:0007204~positive regulation of cytosolic calcium ion concentration |
5 |
3.759398 |
0.017899 |
|
CC |
GO:0005886~plasma membrane |
40 |
30.07519 |
0.027168 |
|
CC |
GO:0070062~extracellular exosome |
36 |
27.06767 |
4.40E-04 |
|
CC |
GO:0005615~extracellular space |
31 |
23.30827 |
1.31E-08 |
|
CC |
GO:0005576~extracellular region |
31 |
23.30827 |
6.59E-07 |
|
CC |
GO:0005887~integral component of plasma membrane |
22 |
16.54135 |
9.33E-04 |
|
CC |
GO:0016324~apical plasma membrane |
12 |
9.022556 |
7.15E-06 |
|
CC |
GO:0048471~perinuclear region of cytoplasm |
11 |
8.270677 |
0.012874 |
|
CC |
GO:0009986~cell surface |
11 |
8.270677 |
0.005205 |
|
CC |
GO:0009897~external side of plasma membrane |
8 |
6.015038 |
8.10E-04 |
|
CC |
GO:0016323~basolateral plasma membrane |
7 |
5.263158 |
0.001783 |
|
MF |
GO:0008270~zinc ion binding |
15 |
11.2782 |
0.045201 |
|
MF |
GO:0005200~structural constituent of cytoskeleton |
5 |
3.759398 |
0.008561 |
|
MF |
GO:0002020~protease binding |
5 |
3.759398 |
0.006364 |
|
MF |
GO:0050699~WW domain binding |
3 |
2.255639 |
0.021335 |
|
MF |
GO:0005496~steroid binding |
3 |
2.255639 |
0.016411 |
|
MF |
GO:0050840~extracellular matrix binding |
3 |
2.255639 |
0.015268 |
|
MF |
GO:0005242~inward rectifier potassium channel activity |
3 |
2.255639 |
0.009183 |
|
MF |
GO:0004111~creatine kinase activity |
3 |
2.255639 |
7.75E-04 |
|
MF |
GO:0004022~alcohol dehydrogenase (NAD) activity |
2 |
1.503759 |
0.049911 |
|
MF |
GO:0004024~alcohol dehydrogenase activity, zinc-dependent |
2 |
1.503759 |
0.042935 |
|
KEGG |
hsa04971: Gastric acid secretion |
7 |
5.263158 |
8.89E-05 |
|
KEGG |
hsa04974: Protein digestion and absorption |
5 |
3.759398 |
0.012074 |
|
KEGG |
hsa04512: ECM-receptor interaction |
5 |
3.759398 |
0.011613 |
|
KEGG |
hsa05204: Chemical carcinogenesis |
4 |
3.007519 |
0.047556 |
|
KEGG |
hsa00980: Metabolism of xenobiotics by cytochrome P450 |
4 |
3.007519 |
0.039157 |
|
KEGG |
hsa04610: Complement and coagulation cascades |
4 |
3.007519 |
0.032797 |
|
Note: BP, Biological process; CC, Cellular component; MF, Molecular function. |
||||
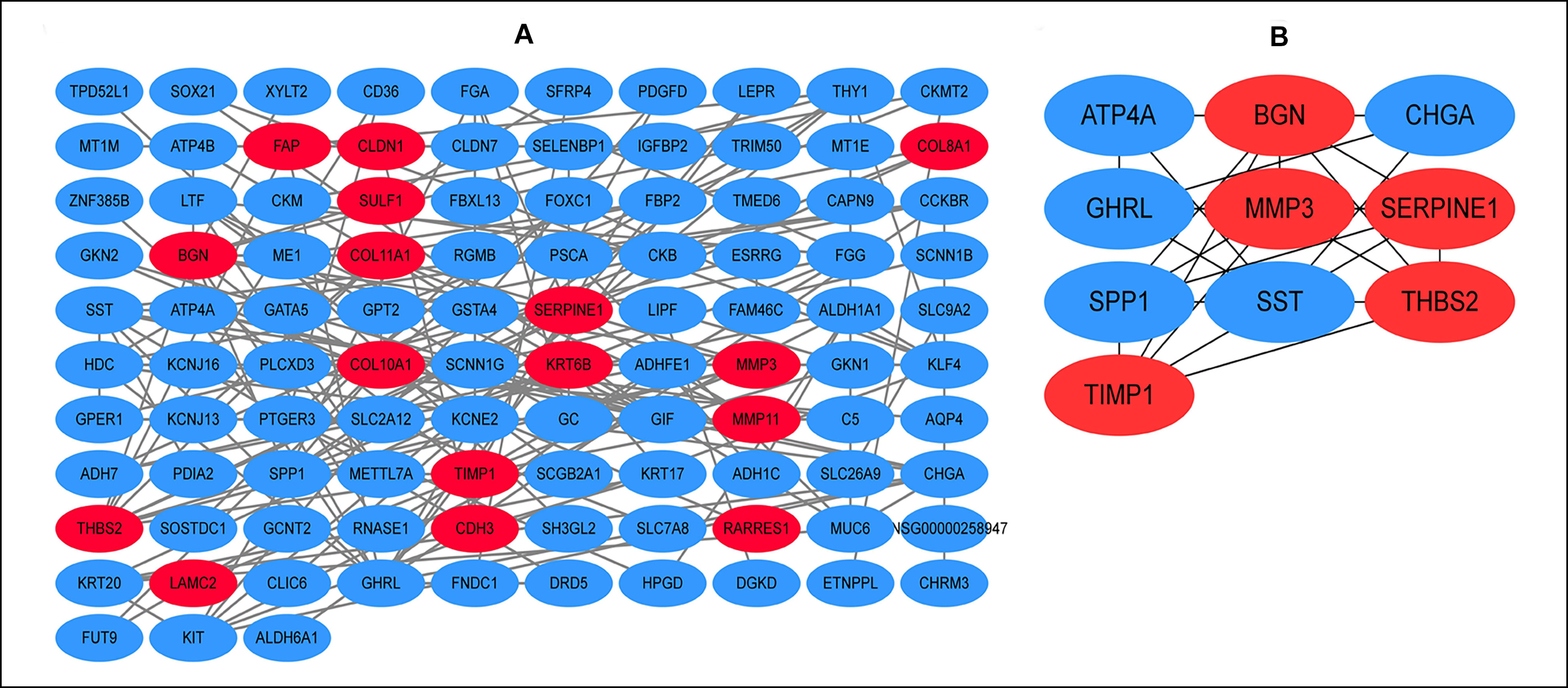 Figure 1: The construction of protein-protein interaction (PPI) network and hub genes analysis. (A) The PPI networks of differentially expressed genes. (B) The top 10 genes in the PPI networks. Red represents upregulated genes while blue represents downregulated genes.
Figure 1: The construction of protein-protein interaction (PPI) network and hub genes analysis. (A) The PPI networks of differentially expressed genes. (B) The top 10 genes in the PPI networks. Red represents upregulated genes while blue represents downregulated genes.
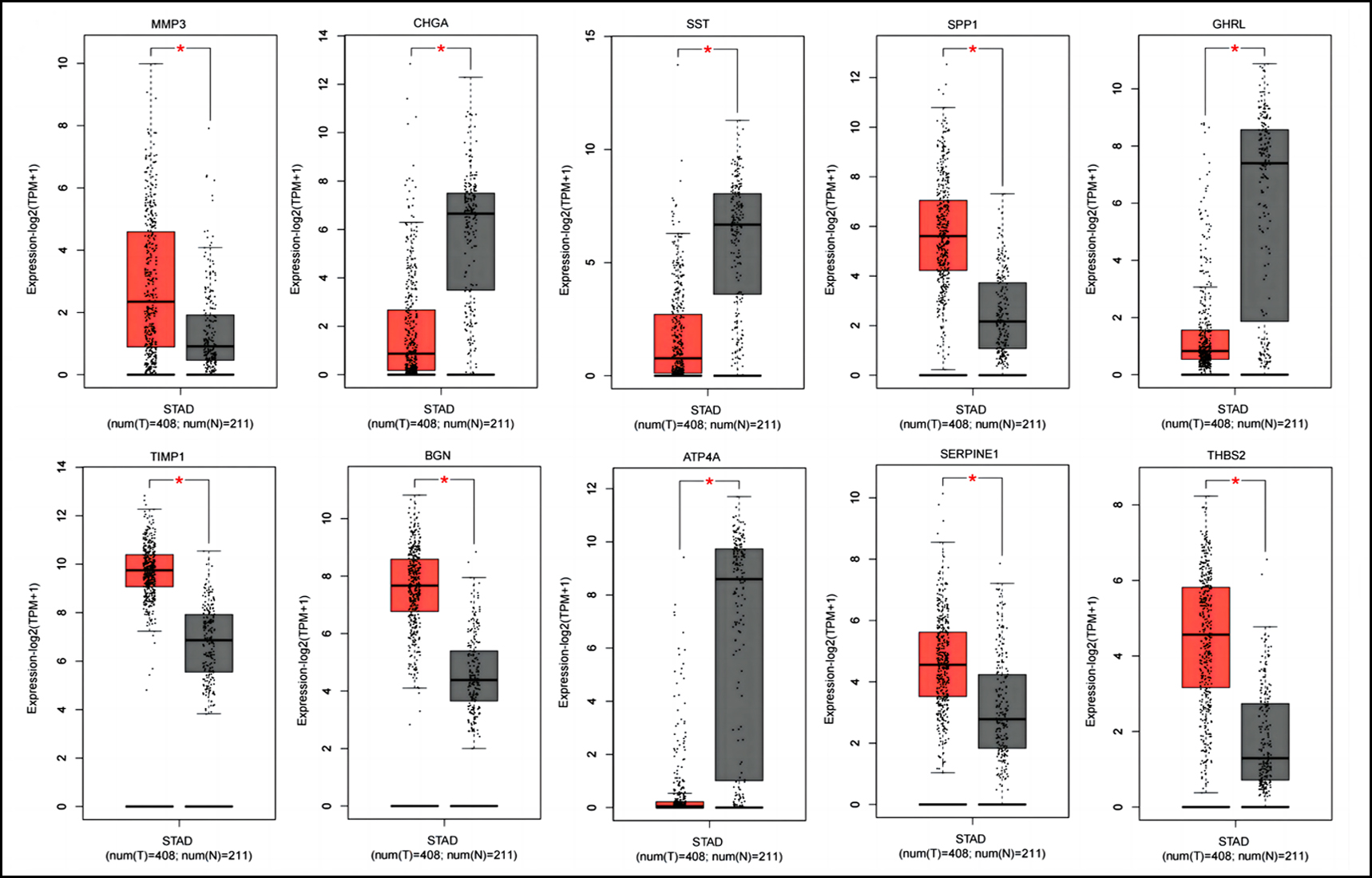 Figure 2(A): Boxplot graphs showing the expression levels of hub genes in tumour and normal tissues of gastric adenocarcinoma patients. *p<0.05, STAD: Stomach adenocarcinoma.
Figure 2(A): Boxplot graphs showing the expression levels of hub genes in tumour and normal tissues of gastric adenocarcinoma patients. *p<0.05, STAD: Stomach adenocarcinoma.
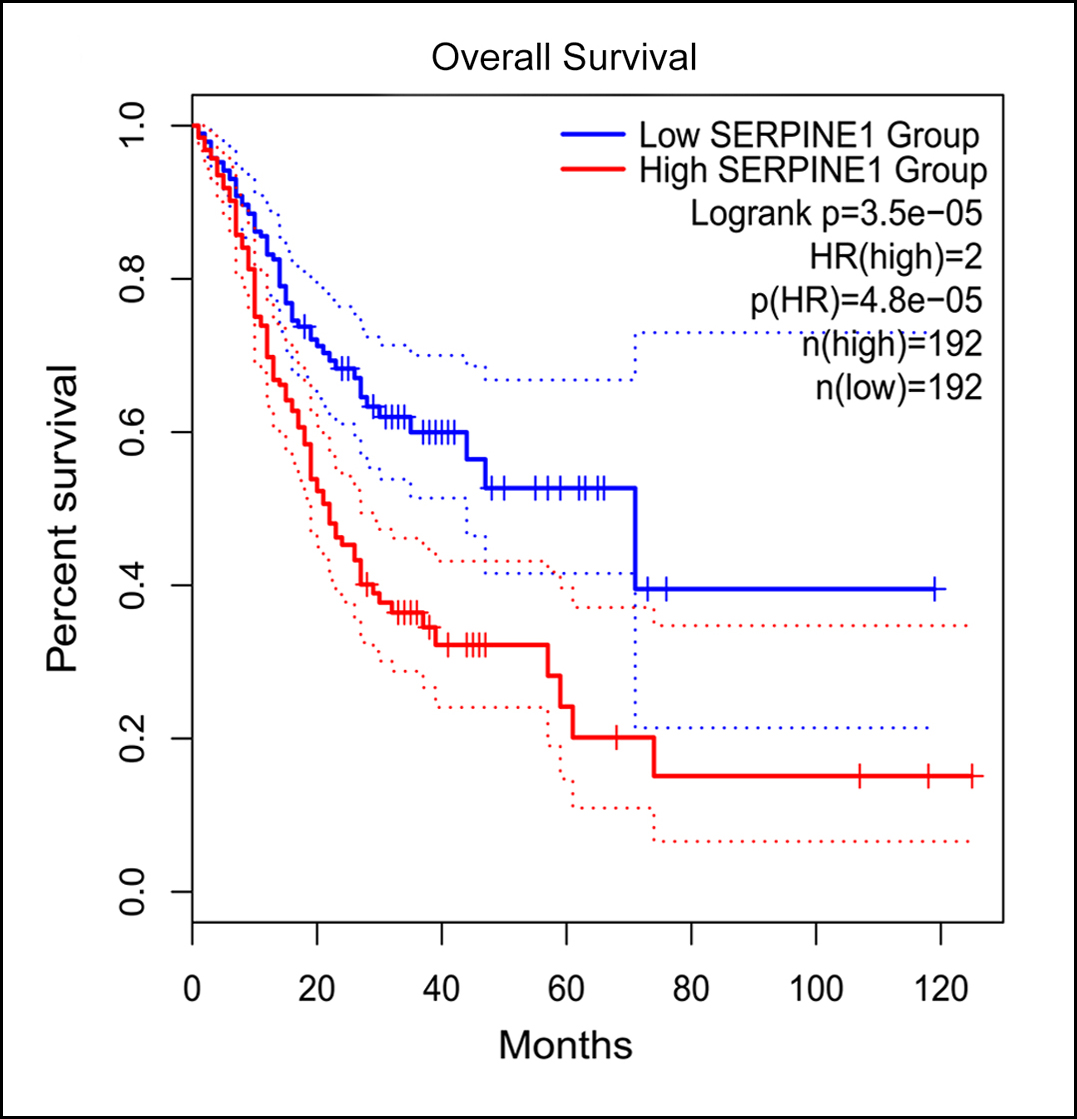 Fiugre 2(B): Kaplan-Meier curves for overall survival in gastric adenocarcinoma patients stratified by SERPINE1 expression level. Cutoff-high and cutoff-low were set at 50% in both groups.
Fiugre 2(B): Kaplan-Meier curves for overall survival in gastric adenocarcinoma patients stratified by SERPINE1 expression level. Cutoff-high and cutoff-low were set at 50% in both groups.
HR: Hazard ratio.
Table II: SERPINE1-targeted medicines.
|
ID |
Name |
Medicine Type |
Medicine Group |
|
DB00029 |
Anistreplase |
Biotech Medicine |
Approved |
|
DB00009 |
Alteplase |
Biotech Medicine |
Approved |
|
DB09130 |
Copper |
Small Molecule Medicine |
Approved |
As shown in Figure 2A, all 10 hub genes (SPP1, SERPINE1, TIMP1, MMP3, BGN, THBS2, SST, GHRL, CHGA, and ATP4A) showed statistically significant expression levels (p <0.05) between gastric adenocarcinoma and normal tissues. According to the GEPIA2 database, the expression trends of 10 hub genes in gastric adenocarcinoma patients and healthy individuals matched those in GEO (Figure 2A). Moreover, compared with high SERPINE1 expression for gastric adenocarcinoma patients, the overall survival rate with lower SERPINE1 expression was significantly higher (Figure 2B), which implied that SERPINE1 might play a pivotal role in the development of gastric adenocarcinoma.
To analyse medicine-gene interaction, SERPINE1 was uploaded into QuartataWeb. Twenty-nine predicted medicines were identified. The top 3 FDA-approved medications were selected, including "Anistreplase", "Alteplase", and "Copper" (Table II). These medicines might be used for gastric adenocarcinoma treatment.
DISCUSSION
Despite significant advances in surgical and medical treatments for gastric adenocarcinoma, its overall mortality rate remains elevated among cancer-related deaths. This high mortality is largely attributed to inadequate early detection, resistance to chemotherapy, and a substantial risk of relapse.2,3 Consequently, there is an urgent need to identify reliable diagnostic and therapeutic biomarkers for gastric adenocarcinoma. With the rapid progression of bioinformatics, a wealth of microarray and sequencing data is now available, facilitating comprehensive and efficient screening of genetic variants and the identification of diagnostic and prognostic molecular mechanisms for various cancers.6,7
In this study, DEGs (1749 in GSE103236, 419 in GSE26942, and 1851 in GSE118916) were identified via GEO2R tools. Upregulation of 30 genes and downregulation of 103 genes were observed. In the GO functional analysis, DEGs predominantly enriched roles related to cell proliferation and migration. These processes are pivotal in the initiation and progression of malignant tumours. Moreover, within the KEGG pathway analysis, DEGs were primarily grouped into ECM-receptor interaction. The interplay between the extracellular matrix (ECM) and its receptors is crucial in the initiation, progression, and metastasis of various cancers.15 In gastric cancer, the ECM constitutes a significant component of the tumour microenvironment. Compared to healthy stomach tissues, the ECM in gastric cancer often exhibits notable remodelling. This transformation correlates with the tumour's invasion, progression, and metastasis.16 Integrins, the primary ECM receptors on the cellular surface, display heightened expression in gastric cancer. This heightened expression is linked to enhanced invasive and metastatic potentials of the tumour. Upon binding to distinct ECM segments, integrins trigger a series of signaling pathways that amplify tumour cell proliferation, migration, and invasion. Metastasis of gastric cancer, especially in regions like lymph nodes and the liver, frequently coincides with modifications in the ECM. These modifications can directly influence the migratory and invasive tendencies of tumour cells.17 In essence, the ECM-receptor interaction is fundamental to the initiation, progression, and metastasis of gastric cancer.
Among the PPI construction, 103 genes/nodes with 160 edges were enriched. Besides, top 10 hub genes (SPP1, SERPINE1, TIMP1, MMP3, BGN, SST, THBS2, GHRL, CHGA, and ATP4A) were selected by the cytoHubba. Then, the authors reconfirmed 10 hub genes through GEPIA2, and the expression trends remained unchanged. Compared to normal tissues, the SERPINE1 was significantly increased in gastric adenocarcinoma tissues. Moreover, compared with high SERPINE1 expression for gastric adenocarcinoma patients, the overall survival rate with lower SERPINE1 expression was significantly higher, which implied that SERPINE1 might play a pivotal role in the progression of gastric adenocarcinoma.
SERPINE1, also termed plasminogen activator inhibitor 1 (PAI-1), is a multifunctional glycoprotein within the serine protease inhibitor superfamily, playing pivotal roles in various cellular processes.18 Recent evidence indicates that SERPINE1 possesses diverse functions in cancer, correlating with adverse outcomes. Enhanced expression of PAI-1 has been linked to increased cell invasion and migration, particularly in esophageal squamous carcinoma.19 Moreover, the lncRNA NKX2-1-AS1 has been identified to promote gastric cancer progression by elevating SERPINE1 expression and activating the VEGFR-2 signalling pathway.20 These studies implied that SERPINE1 might be a potential biomarker of gastric adenocarcinoma.
Targeted medicines can benefit cancer patients through improving treatment efficiency. With SERPINE1 as biomarker, authors identified 3 potential medicines. Anistreplase, a modified congener of streptokinase, has been used in acute myocardial infarction patients since 1990. It transforms fibrin-bound plasminogen into the proteolytic enzyme and plasmin, which degrades fibrin into fibrin degradation products and finally lyses clots.21 Consequently, blood clots and arterial blockages that cause myocardial infarction are eliminated. Alteplase, a recombinant tissue plasminogen activator, is the only medical treatment approved for acute ischaemic stroke.22 Copper, a transition metal and a trace element in the body, is essential for the function of many enzymes including cytochrome-c oxidase and superoxide dismutase.23 According to recent studies, cancer may mediate stroke by inducing coagulation disorders and infections.24 The plasminogen activator system is important for cancer carcinogenesis and invasion, especially carcinomas metastasis.25 Therefore, these medicines may provide new insights in the treatment of gastric adenocarcinoma.
CONCLUSION
SERPINE1 and potential therapeutic agents hold promise to enhance personalised diagnosis and treatment for gastric adenocarcinoma patients in the future. However, the risk of bias in these findings necessitates caution in their interpretation. Future research is essential to validate the utility of these biomarkers.
ETHICAL APPROVAL:
The local ethics committee granted an exemption letter after re-analysing the open-access datasets.
COMPETING INTEREST:
The authors declared no competing interest.
AUTHORS’ CONTRIBUTION:
JL: Study design and manuscript drafting.
JL, NX: Analysed the data and provided interpretations of the results.
WA: Collection and assembling of data.
All authors approved the final version of the manuscript for publication.
REFERENCES
- Yakirevich E, Resnick MB. Pathology of gastric cancer and its precursor lesions. Gastroenterol Clin North Am 2013; 42(2):261-84. doi: 10.1016/j.gtc.2013.01.004.
- Machlowska J, Baj J, Sitarz M, Maciejewski R, Sitarz R. Gastric Cancer: Epidemiology, risk factors, classification, genomic characteristics and treatment strategies. Int J Mol Sci 2020; 21(11):4012. doi: 10.3390/ijms21114012.
- Pasechnikov V, Chukov S, Fedorov E, Kikuste I, Leja M. Gastric cancer: prevention, screening and early diagnosis. World J Gastroenterol 2014; 20(38):13842-62. doi: 10.3748/ wjg.v20.i38.13842.
- Fitzgerald RC, Antoniou AC, Fruk L, Rosenfeld N. The future of early cancer detection. Nat Med 2022; 28(4):666-77. doi: 10.1038/s41591-022-01746-x.
- Hosseini K, Ranjbar M, Pirpour Tazehkand A, Asgharian P, Montazersaheb S, Tarhriz V, et al. Evaluation of exosomal non-coding RNAs in cancer using high-throughput sequencing. J Transl Med 2022; 20(1):30. doi: 10.1186/ s12967-022-03231-y.
- Oliver GR, Hart SN, Klee EW. Bioinformatics for clinical next generation sequencing. Clin Chem 2015; 61(1):124-35. doi: 10.1373/clinchem.2014.224360.
- Chen C, Hou J, Tanner JJ, Cheng J. Bioinformatics methods for mass spectrometry-based proteomics data analysis. Int J Mol Sci 2020; 21(8):2873. doi: 10.3390/ijms21082873.
- Machlowska J, Kapusta P, Baj J, Morsink FHM,Wołkow P, Maciejewski R, et al. High-throughput sequencing of gastric cancer patients: Unravelling genetic predispositions towards an early-onset subtype. Cancers (Basel) 2020; 12(7):1981. doi: 10.3390/cancers12071981.
- Darnet S, Moreira FC, Hamoy IG, Burbano R, Khayat A, Cruz A, et al. High-throughput sequencing of mirnas reveals a tissue signature in gastric cancer and suggests novel potential biomarkers. Bioinform Biol Insights 2015; 9(Suppl 1):1-8. doi: 10.4137/BBI.S23773.
- Dalmer TRA, Clugston RD. Gene ontology enrichment analysis of congenital diaphragmatic hernia-associated genes. Pediatr Res 2019; 85(1):13-9. doi: 10.1038/s41390-018- 0192-8.
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017; 45(D1):D353-61. doi: 10.1093/nar/gkw1092.
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13(11):2498-504. doi: 10.1101/gr.1239303.
- Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014; 8 Suppl (Suppl 4): S11. doi: 10.1186/1752-0509-8-S4-S11.
- Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: An enhan-ced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019; 47(W1): W556-W560. doi: 10.1093/nar/gkz430.
- Bao Y, Wang L, Shi L, Yun F, Liu X, Chen Y, et al. Transcriptome profiling revealed multiple genes and ECM-receptor interaction pathways that may be associated with breast cancer. Cell Mol Biol Lett 2019; 24:38. doi: 10.1186/ s11658-019-0162-0.
- Zhang T, Li X, He Y, Wang Y, Shen J, Wang S, et al. Cancer-associated fibroblasts-derived HAPLN1 promotes tumour invasion through extracellular matrix remodeling in gastric cancer. Gastric Cancer 2022; 25(2):346-359. doi: 10.1007/ s10120-021-01259-5.
- Moreira AM, Pereira J, Melo S, Fernandes MS, Carneiro P, Seruca R, et al. The extracellular matrix: An accomplice in gastric cancer development and progression. Cells 2020; 9(2):394. doi: 10.3390/cells9020394.
- Flevaris P, Vaughan D. The role of plasminogen activator inhibitor type-1 in fibrosis. Semin Thromb Hemost 2017; 43(2): 169-77. doi: 10.1055/s-0036-1586228.
- Wang D, Yang L, Liu Z, Yu J, Zhang M, Zhang Y, et al. PAI-1 overexpression promotes invasion and migration of esophageal squamous carcinoma cells. Yi chuan 2020; 42(3):287-95. doi: 10.16288/j.yczz.19-334.
- Teng F, Zhang J, Chen Y, Shen X, Su C, Guo Y, et al. LncRNA NKX2-1-AS1 promotes tumor progression and angiogenesis via upregulation of SERPINE1 expression and activation of the VEGFR-2 signaling pathway in gastric cancer. Mol Oncol 2021; 15(4):1234-1255. doi: 10.1002/1878-0261.12911.
- Marinac JS, North DS, Stringer KA. Anistreplase: A novel thrombolytic agent for acute myocardial infarction. DICP 1990; 24(6):607-15. doi: 10.1177/106002809002400611.
- Huang X, Cheripelli BK, Lloyd SM, Kalladka D, Moreton FC, Siddiqui A, et al. Alteplase versus tenecteplase for thrombolysis after ischaemic stroke (ATTEST): A phase 2, rando-mised, open-label, blinded endpoint study. Lancet Neurol 2015; 14(4):368-76. doi: 10.1016/S1474-4422(15) 70017-7.
- Percival SS. Copper and immunity. Am J Clin Nutr 1998; 67(5 Suppl):1064S-8S. doi: 10.1093/ajcn/67.5.1064S.
- Dardiotis E, Aloizou AM, Markoula S, Siokas V, Tsarouhas K, Tzanakakis G, et al. Cancer-associated stroke: Pathophysiology, detection and management (Review). Int J Oncol 2019; 54(3):779-96. doi: 10.3892/ijo.2019.4669.
- Abbink K, Zusterzeel PLM, Geurts-Moespot A, van der Steen R, Span PN, Sweep FCGJ. Prognostic significance of VEGF and components of the plasminogen activator system in endometrial cancer. J Cancer Res Clin Oncol 2020; 146(7): 1725-35. doi: 10.1007/s00432-020-03225-7.